Contents
Recent research
Genetic association of thermal limits and transcriptomic thmeral stress response
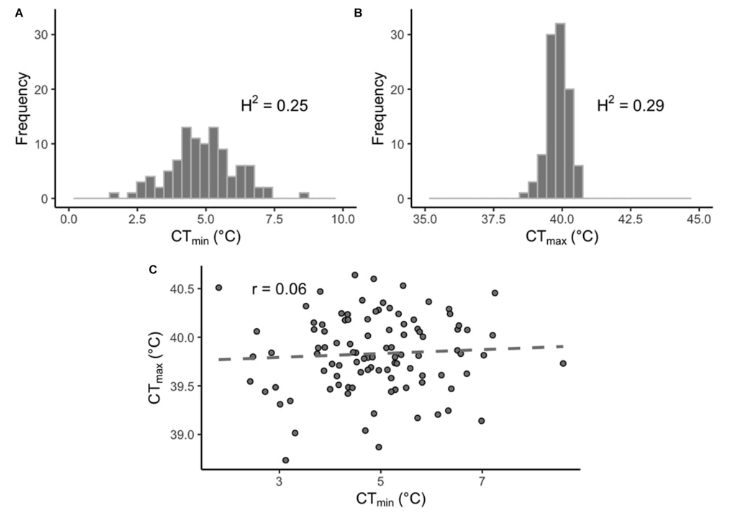
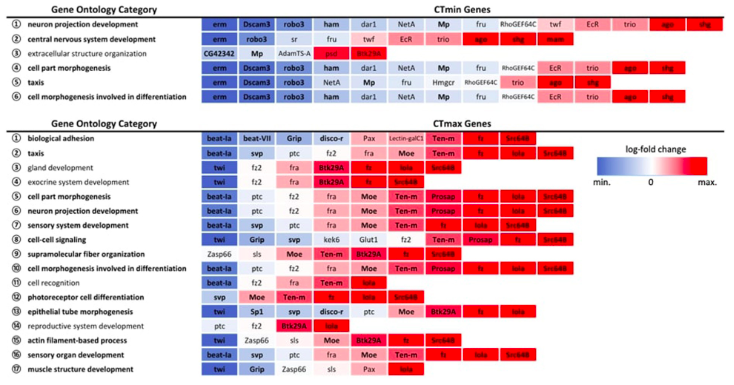
This project aimed to connect the genetic architecture of thermal tolerance in Drosophila melanogaster with gene expression under thermal stress. This work was part of multi-institutional collaborative grant – more information on the grant can be found here. Check out the paper in Frontiers in Genetics here.
Post-undergraduate research
Hypertrophic cardiomyopathy and cMyBP-C
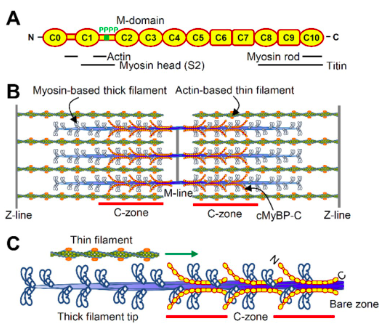
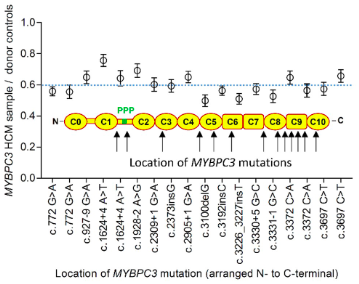
Hypertrophic cardiomyopathy (HCM) is a disease of the heart where the muscle in the ventricle becomes so large that it obstructs the flow of blood. Truncation mutations within the gene MYBPC3, which encodes the protein cardiac myosin-binding protein C (cMyBP-C), are a frequent cause of HCM. This research found that these mutations led to reduced cMyBP-C content in the heart muscle of HCM patients. And that decrease in cMyBP-C content corresponded with in vivo increases the sliding velocity of the thin filament across the C-zone of the thick filament.
Sarcomeric protein turnover
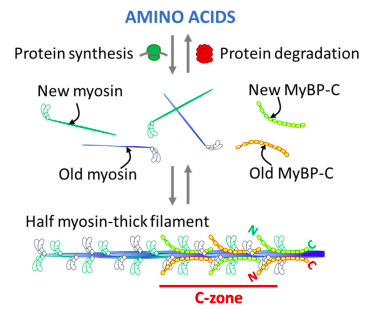
More recent work has investigated the relative turnover rates of sarcomeric proteins in vivo with mice fed a diet of heavy isotope labeling of an amino acid (D3-Leu).
Graduate school course work
Multi-strain vaccine selection on a genotype network
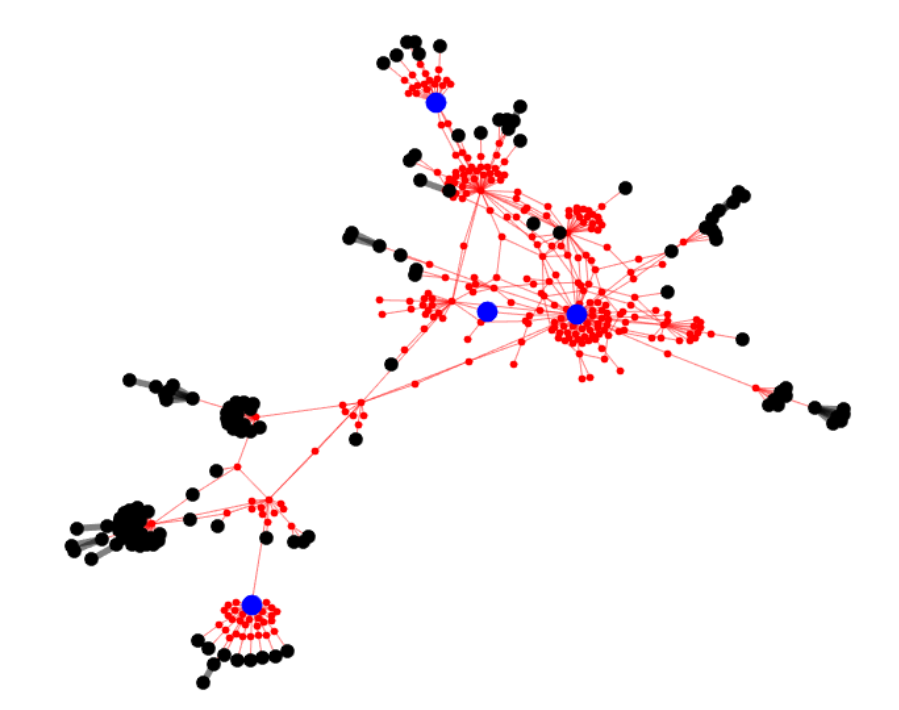
Current strategies to choose strains of a virus for a vaccine involve models of forecasted prevalence without regard to the underlying genotype network of the virus itself.
We used an evolutionary algorithm on an Influenza A genotype network to determine the best strains to select for vaccination. A draft of the paper can be found here. Open source code can be found on GitHub.
Balancing selection
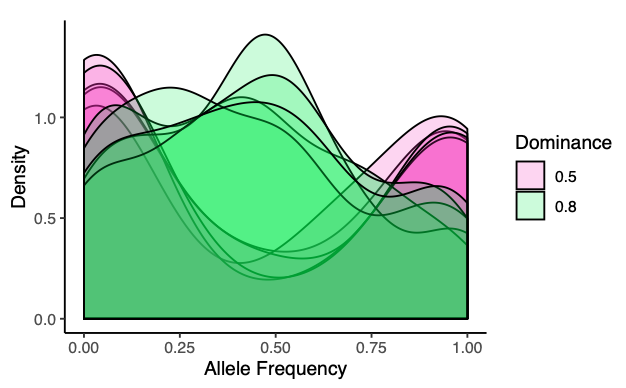
What sort of mechanisms maintain polymorphism within a population? Previous work has shown that seasonal changes in the dominance of an allele can stably maintain polymorphism within a population (Wittmann et al. 2017). We created a similar model to explore the role genetic linkage may play in maintaining polymorphism in this context. A rough draft of our results can be found here. Open source code for the model can be found on GitHub.